MuSIC Project
Browse MuSIC Tools
MuSIC Tools
- This page contains links to the various tools used to construct MuSIC maps. This page will be updated as new MuSIC maps and associated pipelines are released.
Tools for MuSIC map construction
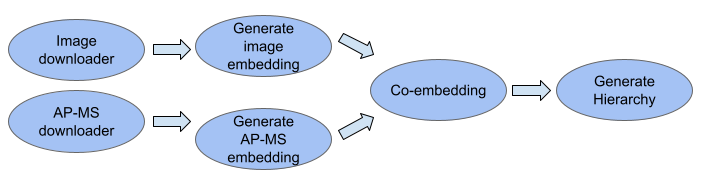
- MuSIC map pipeline
- This package is used to invoke steps of the MuSIC map construction pipeline, including downloading data, generating embeddings, unsupervised co-embedding, and creating the final MuSIC map hierarchy.
- Image downloader
- This package downloads images from the Human Protein Atlas or other user-supplied links
- PPI downloader
- This package loads in a protein-protein interaction network and prepares the data for embedding
- Generate image embedding
- This package creates an image embedding using the densenet model (see Tools for image embedding section below)
- Generate PPI embedding
- This package creates an embedding for each protein in a protein-protein interaction netowrk using node2vec
- Co-embedding
- This package uses unsupervised machine learning to integrate the image and PPI embeddings into a single co-embedding for each protein based on an implementation of MUSE
- Generate Hierarchy
- This package constructs a hierarchy using pan-resolution community detection with HiDeF
- Cell maps utils
- This package contains utilities needed by the various tools
Tools for image embedding
- Image embedding
- This GitHub repository contains code for running the winning model from the Human Protein Atlas Kaggle challenge (Ouyang et al., Nature Methods 2019)
- Single cell embedding
- This GitHub repository contains code for running the winning model from the Human Protein Atlas Single-Cell Classification Kaggle challenge (Le et al., Nature Methods 2022)
Tools for HEK293 MuSIC Map v1.0
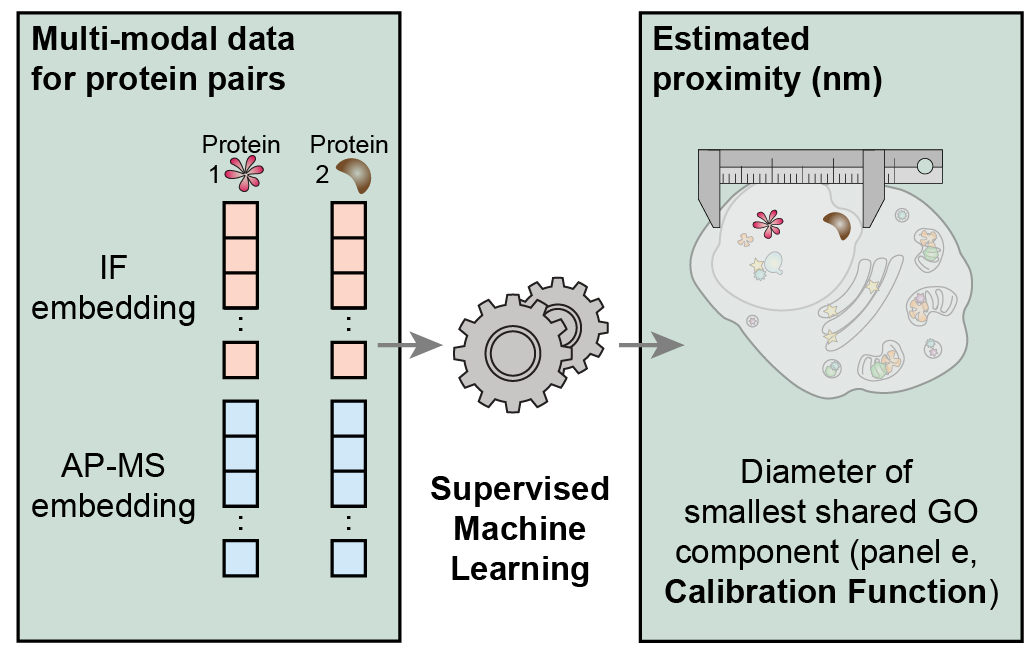
- MuSIC v1.0 map construction
- This GitHub repository contains code and a step-by-step guide for constructing MuSIC maps using the v1.0 pipeline (Qin et al., Nature 2021), which includes supervised machine learning trained with the Gene Ontology.