MuSIC Project
What is the MuSIC project?
The Multi-scale Integrated Cell (MuSIC) project aims to construct the first systematic map of human cell structure and to establish how such maps will transform our ability to treat disease. Towards these goals, we are developing an end-to-end technology pipeline for mapping cellular architecture which incorporates confocal imaging, mass spectrometry and machine learning as key technologies. Thus far, we have demonstrated the feasibility of this pipeline by mapping a small fraction of the cell, covering approximately 3% of proteins (Qin et al., Nature 2021, HEK293 MuSIC Page). We have also begun to integrate cell maps in precision diagnosis of cancers (Zheng et al., Science 2021). We are currently significantly scaling the platform, as well as conducting basic research into technology alternatives, with the goal of achieving a >10X increase in scale and throughput. The ability to efficiently map cell architecture will set the stage for future efforts to use these maps in pharmaceutical and clinical applications and to comprehensively define the nature of human cells across tissues, diseases, and times during development and aging.
Who is part of the MuSIC project?
The MuSIC project is led by the Ideker Lab at the University of California San Diego and the Lundberg Lab at Stanford University, and involves many other collaborators. Members of the MuSIC project team can be found on the Team page.
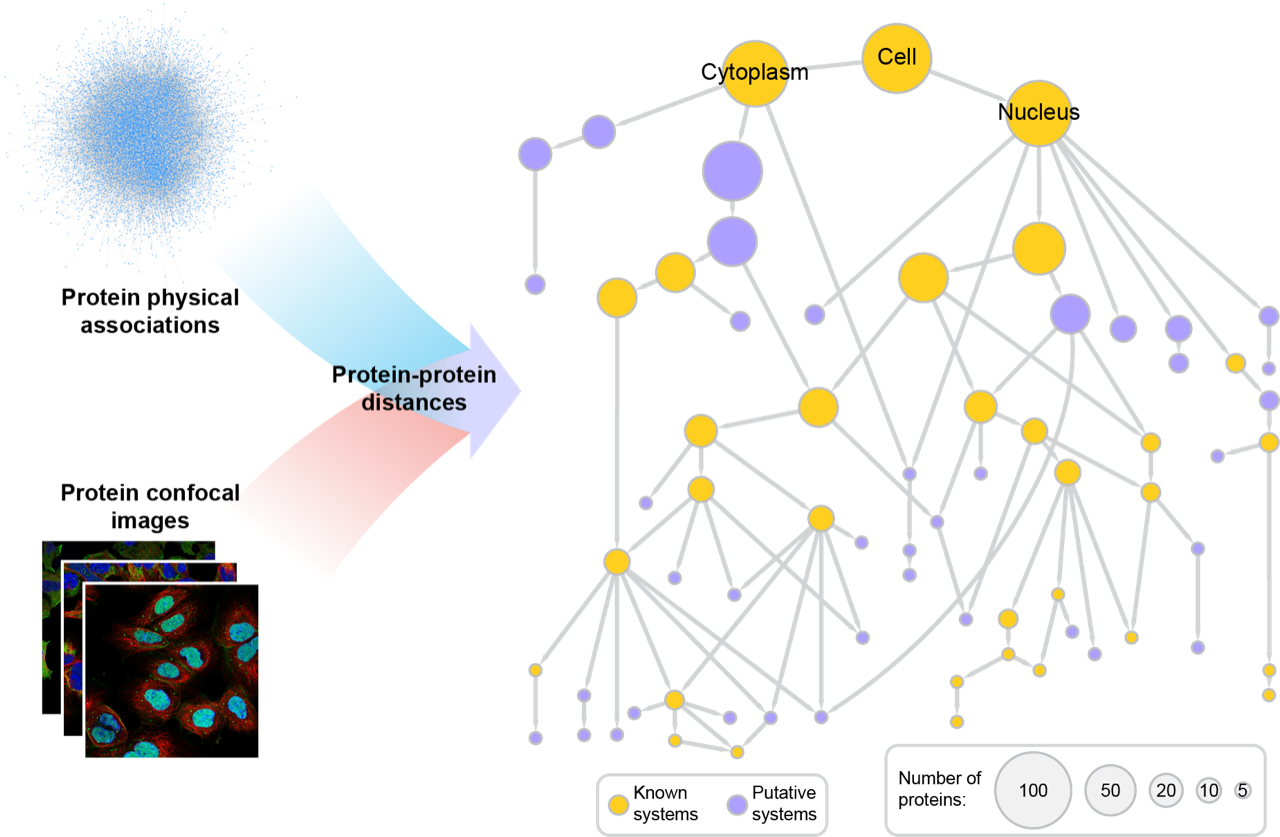
How are MuSIC Maps made?
MuSIC integrates data from numerous datasets, some of which are generated in this project and others in sister projects (e.g., Human Protein Atlas, BioPlex Interactome). Deep neural networks are used to embed each protein in each data modality. For example, node2vec is used to create an embedding for each protein from networks based upon interaction neighborhoods, and Densenet is used to create an embedding for each protein from the imaging data based upon subcellular distributions. Embeddings from the two separate modalities are integrated using either supervised or unsupervised machine learning. Similarities between the integrated embeddings are calibrated to physical distances by calibrating with known subsystems in the Gene Ontology. Pan-resolution community detection is performed to construct a hierarchy of subcellular systems at different scales, from large organelles to small protein complexes. The toolkit to construct MuSIC maps is freely available on GitHub with usage documentation here.
How to cite?
Schaffer, Hu, et al. Multimodal cell maps as a foundation for structural and functional genomics. Nature (2025).
Qin et al. Mapping cell structure across scales by fusing protein images and interactions. Nature 600, 536-542 (2021).